
מונוכרום Shift+A
ניגודיות כהה Shift+S
ניגודיות בהירהShift+D
הגדל גופן Shift+F
הקטן גופן Shift+Z
הדגשת קישורים Shift+X
איפוסShift+C
הצהרת נגישות
© כל הזכויות שמורות 2018







הצהרת נגישות
© כל הזכויות שמורות 2018

MALDI Guided SpatialOMx on the timsTOF fleX
A tumor is more than meets the eye
The microenvironment of cancers contains a combination of healthy cells, tumor cells, connective tissue, blood vessels, and inflammation in different ratios at different time points. Each of these components will have their own unique-molecular signature of compounds. Researchers who study disease states rely heavily on interpreting tissue pathology and creating these maps within the context of biomolecules bridges the gap between traditional OMICS and understanding disease.
SpatialOMx – the key to biomarker discovery
Cancer cells and other disease states have significant genetic and epigenetic modifications that influence the genomic expression cascade. Whether you’re looking at the proteome, lipidome or metabolome, the spatial distribution of compounds contains valuable Information for understanding your sample. If certain compounds are highly spatially concentrated or if molecules are co-distributed on specific structures, this vital information is missing when only examining homogenized samples. OMICS based biomarker discovery can be limited without the benefit of spatial information to add important contextual clues as tissue-level communications networks are integral to cancer growth.
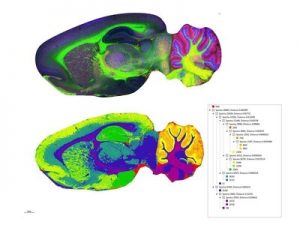
Rat brain lipid image acquired on the timsTOF fleX. Measured at 20 µm spatial resolution, this contains 312,095 spectra with acquisition time of five hours. Lower panel shows segmentation analysis generated from this dataset in SCiLS Lab. The spectra are clustered into regions corresponding to different brain regions.
Bring a spatial dimension to X-Omics analyses
The timsTOF fleX is a versatile instrument for probing the molecular content of your sample. Built on Bruker’s pioneering timsTOF Pro platform, the timsTOF fleX is a fully functional high speed, high sensitivity ESI instrument for all your X-Omics analyses, combined with a high spatial resolution MALDI source and stage specifically designed for resolving molecular distributions and bringing a spatial dimension to OMICS analyses. Transform proteomics analysis into spatial proteomics, lipidomics into spatial lipidomics, and metabolomics into spatial metabolomics.

Create your own Atlases of Intricate Interactions
Mapping complex molecular networks exposes intricate systems of interacting molecules within intracellular networks. Using SpatialOMx to create insightful atlases of communication mechanisms means learning more about the way cells communicate with each other to support change:
how cells interact with the local microenvironment to formulate promotion and mediation processes.
what effect epigenetic modifications have within these networks.
how molecular networks of specific cells interact to perform specific biological functions.
For more information about timsTOF™ Pro